Research
Max Planck Research Group Tessarz
Chromatin is the complex of DNA and proteins that can be found in the nucleus of a cell. We are very interested in understanding how the architecture of chromatin can regulate gene expression. In particular, we aim to elucidate how age-related changes in the epigenome influence transcriptional outputs and ultimately, cellular fate decisions. Over the recent years, we became fascinated by the connection of other cellular pathways with epigenetic mechanisms and have expanded our research to address the connection between metabolism and inter-organellar communication on chromatin architecture. To this end we combine mechanistic approaches in S. cerevisiae and established cell culture systems with analysis of primary cells and tissues using biochemistry, cell biology and a variety of state-of-the art deep sequencing technologies.
Selected projects
Chromatin-mediated regulation of gene expression
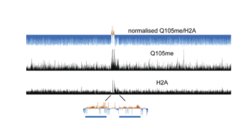
We identified a histone modification (glutamine methylation of histone H2A at glutamine 105) that is exclusively associated with the function of RNA polymerase I. This modification regulates association of rDNA accessibility by inhibiting binding of the histone chaperone FACT, which leads to decreased incorporation of H2A into the rDNA locus (Tessarz et al., 2014). We followed up on this work and identified specific readers of this modification that are connected to ribosome assembly, rRNA processing and sensing of metabolic states. Interestingly, methylation of H2AQ105me depends on another histone modification, acetylation of lysine 56 in histone H3, a modification that has – among others – been associated with rRNA transcription. This line of research also led to the identification of a very interesting cellular crosstalk. Mutations in H3K56 lead to changes in nascent RNA transcription, while steady-state RNA levels are virtually unaffected. Using a genome-wide screen for genetic interactors of H3K56A, we identified the RNA-binding protein Puf5. We could show that Puf5 can either stabilise or degrade transcripts in a context-dependent manner and maintain proper mRNA levels connecting chromatin-mediated regulation of nascent transcription with mRNA degradation pathways (Kochan et al., 2021). Next to our work on histone modification and their role in regulating gene expression, we are also very interested in understanding how chromatin architecture can directly regulate transcription. One example is our work on the histone chaperone FACT and how it regulates access to promoters. In embryonic ES cells, FACT inhibits antisense transcription by maintaining the -1 nucleosome and thus, restrict access to the promoter (Mylonas et al., 2018). Currently, we are extending this work to understand how chromatin architecture and transcription regulation are connected in the context of ageing.
Connection of metabolism and the epigenome
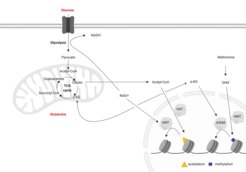
Central metabolites, such as acetyl-CoA, S-adenosylmethionine or the NAD/NADH ratio are intimately linked with epigenetic reactions as they serve as substrates or co-factors for enzymes modifying histones and DNA. Conversely, the induction or repression of metabolic enzymes is under epigenetic control. As ageing is associated with changes in both, metabolism and the epigenome, we are very interested in understanding how the connection between these two essential cellular pathways is impacted. We use mesenchymal stem cells (MSCs) derived from the endosteum of mice to study this interplay. MSCs are important to maintain integrity of bone, but are also essential for haematopoiesis and thus, play a critcal role during the ageing process. From previous work we know that MSCs can differentiate into chondrocytes, osteoblasts and adipocytes to regenerate bone, cartilage and fat in the bone. However, with age, this differentiation is skewed towards the adipogenic lineage. We could demonstrate that these defects in osteogenesis originate from age-associated changes in the chromatin landscape, due to changes in the histone acetylation states in aged MSCs (Pouikli et al., 2020). This change in histone acetylation is a consequence of impaired acetyl-CoA export from mitochondria to the cytoplasm due to lower levels of the carrier (CiC) responsible for this reaction. We uncovered that the decrease in CiC depends on an increase in mitochondrial quality control mechanisms that lead to higher turnover rate of the carrier in aged cells. Strikingly, circumventing acetyl-CoA export by medium supplementation with acetate rescues histone acetylation and osteogenic differentiation capacity of aged MSCs, placing the metabolism-epigenetic interplay at the heart of the differentiation changes. Together with work from other labs, this demonstrates that interventions on the epigenetic level might be a way for cellular rejuvenation.
Epigenetic heterogeneity

One feature of ageing is an increase in biological noise in many tissues. To study this on the level of chromatin and to understand if all cells in our tissues are impacted in the same way, we employ single-cell epigenomic mapping.
In addition, we are very interested in understanding if the potential noise can be maintained in in vitro systems. To study this potential phenomenon of epigenetic memory we use organoid systems that we setup directly from aged tissue.


