Research
Associated Research Group Rorbach
Selected projects
1. Mitochondrial protein synthesis
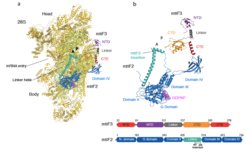
Translation initiation in human mitochondria relies upon specialized mitoribosomes and initiation factors, mtIF2 and mtIF3, which have diverged from their bacterial counterparts. By combining single-particle cryo-EM with fluorescence single-molecule analyses, we have characterised two distinct mitochondrial pre-initiation assembly steps (Khawaja et al. 2020). Our results show that in contrast to what has been postulated for all of the previously studied systems, mtIF3 preserves the assembly of the initiation complex in a way that does not facilitate mRNA binding or accommodation of the initiator tRNA. In addition, our results reveal novel mitochondria-specific molecular interactions between mitoribosomal proteins and initiation factors that were not resolved in the previous reports and postulate the mechanisms of activation of the GTPase action of mtIF2. Finally, supported by single-molecule analysis and biochemical studies, we proposed that translation of mitochondria-specific leaderless mRNA is preferentially initiated on the full monosome, not the small subunit (Remes et al. 2022). As our optimised protocols developed for this project allow for robust purification of mitoribosomes with their associated factors, we are now ready to extend our analyses to describe other stages of translation, focusing on translation termination and recycling.
2. Biogenesis of mitoribosome
Studies of ribosome biogenesis in bacteria and in the eukaryotic cytosol have been in progress for decades, revealing that ribosome production is extraordinarily complex and requires a large number of auxiliary factors. In human mitochondria, only a handful of such factors have been investigated to date. We aim to identify additional assembly factors, as well as structurally and functionally characterise assembly intermediates accumulated upon depletion of the already known assembly factors.
We have initiated characterisation of several GTPases and other auxiliary factors involved in mitoribosome assembly (e.g. GTPBP10, Busch et al. 2019; GTPBP5, Cipullo et al. 2021a). We complement our biochemical analyses with cryoEM, allowing determination of the high-resolution structures of the assembly intermediates (e.g. Cipullo et al.2021b; Itoh et al. 2022). Moreover, we are interested in regulatory mechanisms coordinating mitoribosome assembly with other mitochondrial processes according to the cellular and environmental demands.
3. Mitochondrial RNA binding proteins
In all biological systems, mRNAs are associated with RNA-binding proteins (RBPs), forming complexes that control gene regulation, from mRNA synthesis to decay. The post-transcriptional regulation in mammalian mitochondria is conducted by several mitochondrial RBPs (mt-RBPs) at various stages of mt-RNA metabolism.
We aim to identify novel mitochondrial factors involved in RNA metabolism and translation by means of biochemical, proteomic and genetic screens, followed by the careful characterisation of phenotypes associated with aberrant expression of these genes. For example, our study identified C6orf203 as a novel RNA-binding protein interacting with the mitoribosomal large subunit (Gopalakrishna et al. 2019). We complement our biochemical characterizations with structural studies.
4. Mitochondrial gene expression in disease
We expanded our mechanistic studies of mitochondrial gene expression into primary cells from mitochondrial disease patients. Several novel candidate genes with pathogenic mutations have recently been identified using whole exome sequencing in the diagnostic centres that collaborate with us. We perform detailed molecular analysis of cell lines derived from such patient material to understand how the pathological mutations affect mitochondrial function and lead to disease.
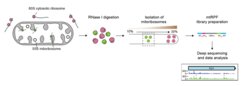
To investigate the function of the factors involved in mitochondrial diseases, we employ, for example, the emerging technology of ribosome profiling, which maps the exact positions of ribosomes on transcripts by nuclease foot-printing. Deep sequencing of the nuclease-protected fragments reveals the exact region of the transcriptome currently being translated, with the abundance of different footprint fragments reporting the amount of translation. We have already successfully applied this technique towards understanding the mechanisms of action of several mitochondrial factors (Pearce et al. 2017, Gopalakrishna et al. 2019, Krüger et al. 2022) and adapted the method for studies of patient-derived samples.