Research
Research Group Matić

ADP-ribosylation (ADPr) is a versatile post translational modification (PTM) that plays critical roles in many physiological and pathological processes, from the DNA damage response (DDR) and cancer to neurological disorders and ageing. ADP-ribose is added to different amino acids on target proteins by the action of various transferases, most notably PARPs and sirtuins. Both families of enzymes use NAD+ as substrate, thereby regulating NAD+ metabolism. PARP1, the main DNA-dependent ADP-ribosyltransferase and an important target for cancer therapy, has been studied extensively in the DDR, with functions in multiple DNA repair pathways. Upon activation, PARP1 primarily ADP-ribosylates itself and histones, which promotes the recruitment of DNA repair factors to chromatin. As genome instability caused by persistent DNA damage together with altered epigenetic landscape have been established as mechanisms driving organismal ageing, the role of PARP1 in ageing processes has attracted considerable attention especially in the context of sirtuins and NAD+ metabolism. Despite a wealth of evidence pointing to the biomedical importance of ADP-ribosylation signalling, especially in DDR, cancer therapy and ageing, the molecular basis by which PARP1 and sirtuins exert their functions has been difficult to determine. In particular, little has been determined about the specific roles of histone ADPr, in large part due to the lack of biochemical tools and knowledge of the underlying molecular mechanisms.
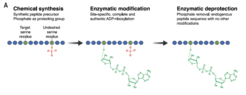
Owing to its chemical instability, studying ADPr at the molecular level is very challenging. Using advanced proteomics, we revealed Serine ADPr as a new type of PTM, targeting all histones upon DNA damage (Leidecker et al. Nature Chemical Biology 2016). Strikingly, this important cellular signal had eluded discovery for half a century, although it has since been demonstrated to be a major PTM that targets thousands of modification sites. This pioneering identification of Ser-ADPr set the stage for our discovery that HPF1 switches PARP1’s specificity to modify serines on itself, histones and many other proteins (Bonfiglio et al. Molecular Cell 2017). Shortly after this identification of HPF1-PARP1/2 complex as the ‘writer’ of Ser-ADPr, we elucidated ARH3 as its ‘eraser’ (Fontana et al. eLife 2017) and showed that Ser-ADPr is the major form of ADPr in DDR (Palazzo et al. eLife 2018).
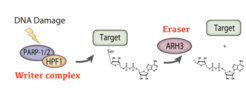
Since 2020, Ser-ADPr by HPF1/PARP1 has become an intensely studied topic with a clear relevance to the development of anti-cancer drugs. Despite this strong and growing interest in Ser-ADPr, further progress was constrained by a lack of biochemical tools and approaches. To address this issue, we turned our discovery of Ser-ADPr by PARP1/HPF1 into a powerful chemical biology approach that uses a competing PTM to “steer” the enzymatic reaction (Bonfiglio et al. Cell 2020). This phopho-guided strategy enabled us to generate the first-ever site-specific ADPr antibodies as well as sensitive broadband mono-ADPr-specific antibodies. Thanks to these novel tools, we made another highly unexpected discovery: upon DNA damage PARP1 targets, most notably histones, are predominately mono-ADP-ribosylated. While it was assumed for decades that the only output of PARP1 signaling is poly-ADPr, we showed that the poly-ADPr hydrolase PARG trims the transient polymer back to mono-ADPr, which then remains as the predominant species in the DNA damage response.
By applying SpyTag technology, we can now detect even low levels of ADPr in a variety of applications, even following modification events in real time in living cells (Longarini et al. Mol Cell 2023). Armed with these new tools, we showed that chromatin (primarily histone) mono-ADPr constitutes the second wave of PARP1 signaling in the DNA damage response and that HPF1 converts PARP1 into a mono-ADP-ribosyltransferase in cells. To validate the concept of PARP1-catalysed mono-ADPr as an important carrier of biological information, we combined three complementary proteomics approaches to identify a set of intriguing readers of histone mono-ADPr. Focusing on one of these readers, the ubiquitin ligase RNF114, we demonstrated that Serine mono-ADPr functions as a recruitment signal to modulate the DNA damage response and telomere maintenance.
Building upon these seminal discoveries and combining our advanced proteomic/chemical biology strategies with traditional biochemical and cellular approaches, we aim to deepen our functional and mechanistic understanding of how ADPr regulates DNA repair and the process of ageing. Currently, based on the surprising prevalence of mono-ADPr upon DNA damage, we are testing the hypothesis that Ser-mono-ADPr regulates the recruitment of proteins to the sites of DNA damage. Recruitment of DNA repair factors to chromatin has long been recognized as one of the main functional outcomes of PARP1-dependent signaling pathways. Yet it has always been attributed to poly-ADPr, while mono-ADPr was neglected, being effectively “invisible” for decades. To determine the functional consequences of Serine mono-ADPr, we now aim to identify and characterize the ‘readers’ of histone mono-ADPr by applying state-of-the-art quantitative mass spectrometry and broadening the scope of our chemical biology strategy.
Overall, our research aims to dissect the molecular mechanisms by which histone ADPr regulates ageing-related biological and disease processes, focusing on specific Ser-ADPr marks and the new possibility of mono-ADPr as a key regulator of DNA repair.