Research
Max Planck Research Group Huppertz
While transcriptional changes have received scholarly attention for many decades, RNA-binding proteins (RBPs) have only recently taken centre stage as regulators of metabolism. RBPs are a versatile group of proteins that can facilitate short- and long-term metabolic adjustments of cells undergoing cell division and differentiation. Furthermore, RBPs can integrate metabolic stimuli through, for example, post-translational modifications, changes in localization or metabolite availability. Our overarching aim is to utilize the metabolically dynamic system of quiescent and activated NSCs during ageing to elucidate the following questions and discover the involvement of canonical and non-canonical RBPs therein:
- What regulates the metabolic alterations that ageing NSCs undergo?
- What role does metabolism play in the ageing process of NSCs?
- What coordinates cytosolic and mitochondrial metabolic pathways in ageing NSCs?
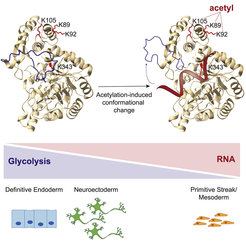
One example of a canonical RBP that impacts the metabolic setup of the cell is YBX3. By binding to the 3’ untranslated region, YBX3 stabilizes the transcript of the amino acid transporter SLC7A5, and thereby indirectly alters the availability of large, neutral amino acids in the cell (Cooke et al., 2019). In addition, many essential metabolic enzymes have been identified to bind RNA in different cell types and organisms. Two simplistic modes of RNA–enzyme interactions can be envisioned. On the one hand, metabolic enzymes might moonlight as RBPs and regulate the fate of their target RNAs. On the other hand, RNA might regulate these enzymes, a process we have recently described for the glycolytic enzyme Enolase (Huppertz et al., 2022; Figure 1). This very large class of non-canonical RBPs might form a novel layer of metabolic regulation.
Our aim is the development of a combinatorial tool for the metabolism-centric classification of ageing NSCs using genetically encoded FRET-based sensors for key metabolites. The powerful RNA interactome-capture method (Perez-Perri et al., 2018) will enable the global identification of proteins differentially associated with RNA in cells with distinct metabolic profiles as measured by our sensors. Candidate proteins with dramatically changed RNA-binding potential will be knocked out, where possible, using CRISPR/Cas9 and the cells will be subjected to transcriptomic and metabolic assessment. The results of this in-depth cellular characterization will be integrated with CLIP datasets of candidate RBPs to understand their role in establishing the metabolic profile of ageing NSCs.
Metabolism affects the acetylation and methylation of histones and in turn alters the RNA expression landscape of ageing NSCs. Therefore, it is essential to connect the metabolic state and global RNA expression profiles at the single-cell level. Single-cell transcriptome sequencing will be used on the FRET sensor-expressing cells to characterize RNA expression levels and directly link it to the metabolic state of the cell. This will enable temporal tracing of NSCs while simultaneously assessing metabolic gradients, potentially revealing heterogeneously expressed markers that are relevant for priming cells for a specific age-related fate.
Working on Enolase 1, a non-canonical RBP, has uncovered a novel world of metabolic regulation that has the potential to be universal. Therefore, we will dedicate part of our research to the study of non-canonical RBPs like the mitochondrial tricarboxylic acid cycle enzymes and the role their RNA binding might play in coordinating metabolic alterations as part of the ageing process.
