Max Planck Research Group Demetriades
Research
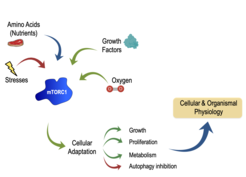
The mechanistic target of rapamycin (mTOR)—primarily as part of mTOR complex 1 (mTORC1)—is a master regulator of growth, metabolism, secretion, and autophagy. Underscoring the importance of mTORC1 for the maintenance of cellular homeostasis, a large number of inputs converge on this complex to regulate growth. Nutrients, energy and growth factors activate mTORC1, whereas various stresses strongly inhibit its activity (Fig. 1). Importantly, mTORC1 does not function as an "ON-OFF switch", but rather as a finely tuned rheostat: quantitative fluctuations of mTORC1 activity determine the cellular metabolic and, thus, growth program. The immediate upstream activator of mTORC1 is the small GTPase Rheb, whose activity is in turn regulated by the inhibitory Tuberous Sclerosis Complex (TSC). Hyperactivation of mTORC1—caused mainly by mutations on its upstream regulators like the TSC—is of clinical relevance and a common feature of most cancer types. Moreover, perturbations of mTOR activity are linked to ageing, lifespan, and healthspan. Accordingly, the immune system decline, muscular atrophy, as well as the onset and progression of multiple age-related, metabolic (diabetes) and neurological disorders (cognitive decline, Alzheimer's disease), are—at least in part—caused by dysregulated mTOR function. Consequently, a number of pharmacological inhibitors that target components of the mTOR network are currently applied in the clinic for the treatment of mTOR-related diseases.
Our lab investigates how cells control their growth in health and disease. In particular, our projects aim to understand:
- How is cell growth regulated in normal cells?
- How does its dysregulation contribute to the development of age-related disease?
- How do cells sense the presence or absence of nutrients, to regulate growth?
- How is information from multiple diverse signals integrated to regulate TSC/mTORC1?
- How are these protein complexes regulated in a spatiotemporal and tissue-specific manner?
Unravelling the AA-sensing regulatory network upstream of mTORC1
The availability of nutrients, such as amino acids (AAs), is a prerequisite for cell growth, hence a robust mTORC1 regulator. Previous studies on how AAs regulate mTORC1 have mainly focused on the lysosomal Rag GTPases and built a complex protein network that coordinately senses AAs to modify Rag activity. According to the current model, AA sufficiency leads to Rag activation, which in turn recruits mTORC1 to the lysosomal surface, where its direct activator Rheb also resides. Importantly, several aspects of the spatiotemporal mTORC1 regulation by AAs remain so far elusive. Therefore, many of our projects aim to understand how nutrient availability signals to drive growth and metabolism via the TSC/mTORC1 signaling axis. We study in detail both the lysosomal Rag-dependent AA-sensing machinery, and the Rag-independent mechanisms via which AAs regulate mTORC1. We have recently explored the evolution of the lysosomal AA-sensing machinery from yeast to humans and characterized the importance of the structural and functional diversification of the Rag paralogous genes in mammals [see Gollwitzer et al., 2021 Nat Cell Biol]. Moreover, we investigate the cross-talk between different nutrient sources (amino acids, lipids, sugars) in the regulation of mTORC1 [see Nicastro, Brohée et al., 2021 bioRxiv]. Collectively, this work identifies new mechanisms and principles of mTORC1 activation and thus expands our view on how AAs and other nutrients control mTORC1 activity. In addition, it provides novel mTORC1 regulators, as putative targets for drug development against mTOR-related diseases.
Signal integration and lysosomal relocalization of the TSC complex
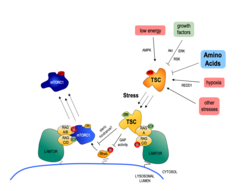
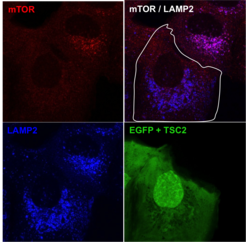
The main upstream negative regulator of mTORC1 is the heterotrimeric TSC protein complex. Notably, virtually all upstream stimuli that regulate mTORC1, including AAs, signal at least in part via the TSC [see Demetriades et al., 2016 Nat Commun]. Therefore, the TSC/mTORC1 hub functions as an integration point for stress signaling and coordinates the cellular stress response (Fig. 2).
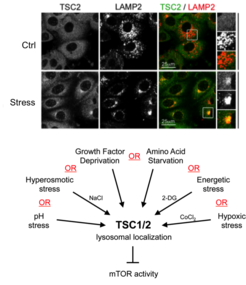
Our previous work revealed the cellular and molecular mechanisms by which mTORC1 is inactivated in response to amino acid starvation. When cells lack amino acids, the TSC is rapidly recruited to the lysosomal surface by the Rag GTPases to act on its target Rheb and, thus, influence mTORC1 localization and activity. This work placed TSC in the AA sensing pathway and showed that AA starvation inactivates mTORC1 via changes in the subcellular localization of TSC [see Demetriades et al., 2014 Cell] (Fig. 3).
In a follow-up study, we revealed that the lysosomal relocalization of TSC is a universal response to cellular stress; each individual stress stimulus, when applied singly to cells, is sufficient to drive lysosomal recruitment of TSC, thereby inhibiting mTORC1 [see Demetriades et al., 2016 Nat Commun]. Hence, the Boolean Operator for the lysosomal relocalization of TSC in response to multiple stimuli is the "OR" operator (Fig. 4). This way, cells ensure that, under unfavorable conditions, mTORC1 will become inactive to cease growth, thus preventing a metabolic catastrophe and, ultimately, cellular death. One important but poorly understood stress stimulus is hyperosmotic stress. Consequently, we also focused on the signaling events by which osmostress inactivates mTORC1 and put together the complete signaling pathway, which involves multiple kinases that impinge on TSC2 and regulate its localization [see Demetriades et al., 2015 Sci Rep]. Our previous work, together with the Teleman lab, also highlighted the role of Tsc2 and the spatial distribution of TORC1 activity in a developing organ in Drosophila, and suggested a role of this machinery in wound healing [see Romero-Pozuelo et al., 2017 Dev Cell]. More recently, together with our collaborators, we discovered two novel factors that control the TSC lysosomal recruitment and mTORC1 inactivation: the stress-granule-related G3BP1/2 proteins that form interactions with TSC2 [see Prentzell et al., 2021 Cell]; and the lysosomal PIP2 phospholipids that anchor the TSC complex via TSC1 (see Fitzian et al., 2021 Mol Cell]. Taken together, these projects revealed the qualitative and quantitative aspects of how multiple upstream stimuli are mechanistically integrated to regulate cell growth in a spatiotemporal manner.
mTOR controls secretory pathways to reshape the extracellular proteome
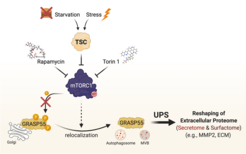
We recently highlighted the TSC/mTORC1 hub as a master controller of unconventional protein secretion (UPS) via the phosphorylation of GRASP55 at the Golgi [see Nüchel et al., 2021 Mol Cell] (Fig. 5). We revealed how cells reshape their extracellular proteome to adapt to multiple cellular stress stimuli, thus underscoring the biological role of UPS. Of note, we identified mTORC1 as the first signaling pathway that regulates UPS, thereby linking physiological nutrient/stress signaling to the cellular stress response; and GRASP55 as a direct, Golgi-based effector of mTORC1, thus revealing a physiological role for mTORC1 at this organelle. Finally, we provided important mechanistic insights into the cell biology of UPS, and demonstrated how mTORC1 and GRASP5 alter the secretome and surfactome upon stress.
Our current research follows up on our previous findings and aims to shed light on the molecular mechanisms of cellular growth control and the regulation of the TSC/mTOR axis. It centers around the fields of mTOR signaling, nutrient sensing, TSC biology, and secretion/ECM biology; and, therefore, it covers important aspects of both basic and translational research. Ultimately, our goal is to add novel targets for drug development to the arsenal against mTOR-opathies and to tackle ageing.




