Research
Max Planck Research Group Panier
RNA-dependent DNA damage responses
Our first research focus area centers around a recent paradigm shift in the genome stability field which is the realization that the events that occur on damaged chromatin to repair DNA lesions are dependent on RNA and RNA-binding proteins.
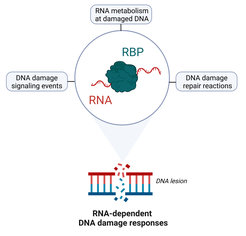
While many RNA-binding proteins have well-characterized roles in adjusting gene expression in response to genotoxic stress, it is becoming increasingly clear that these proteins also have non-canonical functions in the DNA damage response that go well beyond transcription, splicing and mRNA processing (Figure 1). We use a wide variety of cell biological and omics approaches to understand
- How RNA-binding proteins help to process, stabilize and regulate RNAs in the context of damaged chromatin at bulky DNA lesions (i.e. nucleotide excision repair) and at DNA double-strand breaks.
- How this facilitates DNA damage sensing, signaling and repair of these types of lesions.
- Whether and how the dysfunction of specific RNA-binding proteins promotes genome instability and cellular senescence in a physiological context, for example in neurons.
Telomeres
Our second research focus area investigates how cellular machineries that deal with replicative stress and repair of DNA double-strand breaks crosstalk with the mechanisms that ensure telomere stability and function.
Telomeres are critical for genome stability as they protect chromosome ends from degradation and fusion by cellular DNA repair processes. In non-malignant somatic cells, telomeres undergo progressive shortening after DNA replication, which eventually results in replicative senescence and checkpoint-driven cell death, a phenomenon, which plays a key role in the ageing of the organism. In contrast, tumour cells are able to counteract telomere attrition, and thus achieve replicative immortality, by either re-expressing telomerase or inducing alternative lengthening of telomeres (ALT), which relies on telomere recombination.

We recently identified the protein SLX4IP as a novel regulator of genome maintenance in human cells. We found that SLX4IP is enriched specifically at ALT telomeres where it is critical to the control of the DNA damage response machinery that promotes telomere recombination (Panier et al. Mol Cell 2019) (Figure 2).
We now aim to understand:
- How SLX4IP interacts with the replication stress response machinery that is hyperactived at ALT telomeres (Figure 2)
- Whether and how this protein and its associated factors ensure genome stability also at non-telomeric loci and in non-ALT cells
- How their dysfunction may contribute to genome instability-triggered and ageing-associated diseases

